What is OMERO?
OMERO is a software platform for managing, sharing and analysing images data. OMERO supports over proprietary 150 file formats, including all major microscopy formats, medical images, digital pathology images, high content screening, etc., using Bio-Formats. Bio-Formats is a Java software tool for reading proprietary image data and metadata and writing image data using standardized open formats.
OMERO handles all your images in a secure central repository. Users can view, organize, analyze and share data from anywhere via the internet. Users can work with image data and metadata from a Desktop application, from the Web or from 3rd party tools e.g. Fiji.
OMERO stores image metadata in a relational database and offers a more flexible structure based on HDF5 to store for example, analytical results. This allows analytical results generated by 3rd party softwares e.g. CellProfiler, ilastik, etc., to be stored alongside the images.
Recommendations and software tools are being developed to capture acquisition metadata importable into OMERO e.g. 4DN-BINA-OME-QUAREP (NBO-Q).
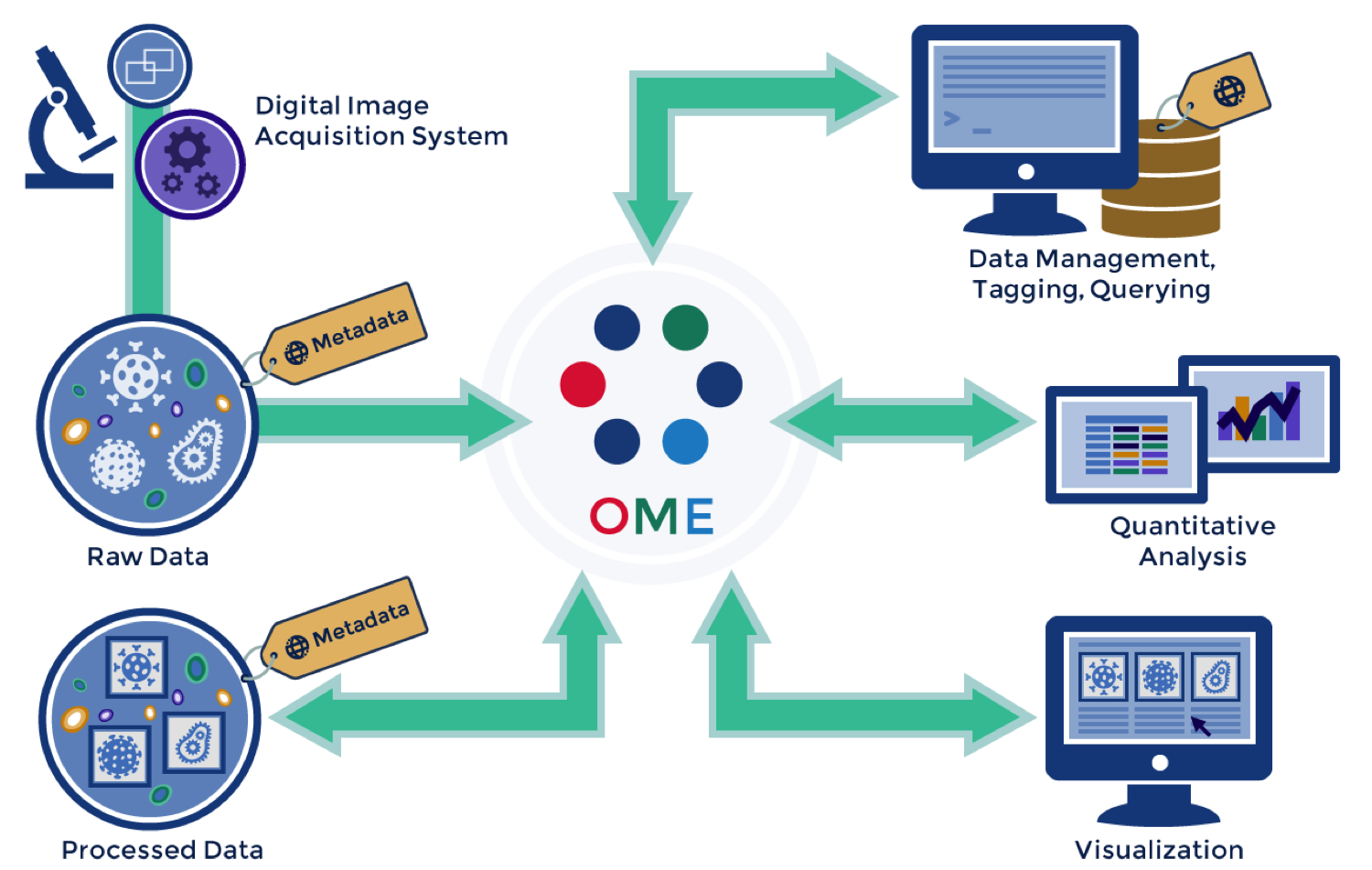
Schematic overview of the OMERO tool assembly.
Who is OMERO intended for?
OMERO is designed to be an institutional repository. It offers a secure central way for scientists, researchers and data stewards to handle their imaging data. All the image data from a facility can be securely stored and managed, using group permissions and user roles to allow controlled access tailored to your institution. From private repositories for sensitive data to hosting public data for your website and latest publications, the permissions model is designed to meet the range of researchers’ needs. OMERO is tried and tested in hundreds of institutions world-wide, with extensive installation and configuration documentation for system administrators and community support via dedicated mailing lists and forums.
The OMERO platform uses a Group/User permission system. The degree to which their data is available to other members of the group depends on the permissions settings for that group. Whenever a user logs on to an OMERO server, they are connected under one of their groups. All data they import and any work that is done is assigned to the current group, however the user can move their data into another group. Users require login credentials to access the system. OMERO also supports the use of an LDAP server.
OMERO is an open-source system designed to be extended and integrated with other tools. The OMERO API allows clients to be written in Java, Python, C++ or MATLAB. The OMERO platform includes a Java client OMERO.insight, a Python-based web client OMERO.web, a Command Line Interface which uses Python, and a Java ImageJ plugin. OMERO also supports a scripting service which allows Python scripts to be run on the server and called from any of the other clients. A vast amount of documentation and code examples for scientists, developers and system administrators are available.
A demo server maintained by the OME team is also available for users wishing to try out, request an account.

3rd party tools and OMERO
Which tasks can be solved with OMERO?
OMERO can be used for the day-to-day data management of data.
- Users can remotely view, handle metadata, analyze, generate figures ready for publication, etc.
- The plaform can also be used to publish data either using public repository like Image Data Repository (IDR) or by enabling the public user within the OMERO installation in a given institution e.g. Liverpool CCI gallery.
- It is the software platform for several public image repositories e.g. Image Data Resource (IDR),EMPIAR
- It is also used as a teaching platform by several institutions e.g. University of Dundee, Harvard Medical school.
Related pages
More information
Training
Tools and resources on this page
| Tool or resource | Description | Related pages | Registry |
|---|---|---|---|
| 4DN-BINA-OME-QUAREP (NBO-Q) | Rigorous record-keeping and quality control are required to ensure the quality, reproducibility and value of imaging data. The 4DN Initiative and BINA have published light Microscopy Metadata Specifications that extend the OME Data Model, scale with experimental intent and complexity, and make it possible for scientists to create comprehensive records of imaging experiments. The Microscopy Metadata Specifications have been adopted by QUAREP-LiMi and are being revised in QUAREP-LiMi in collaboration with instrument manufacturers | Bioimaging data | Standards/Databases |
| Bio-Formats | Bio-Formats is a software tool for reading and writing image data using standardized, open formats. | Tool info | |
| CellProfiler | Image analysis software | Tool info Training | |
| EMPIAR | Electron Microscopy Public Image Archive is a public resource for raw, 2D electron microscopy images. You can browse, upload and download the raw images used to build a 3D structure | Bioimaging data Structural Bioinformatics Data publication | Tool info Standards/Databases Training |
| Fiji | Fiji is an image processing package | Tool info Training | |
| ilastik | ilastik is a user-friendly tool for interactive image classification, segmentation and analysis | Tool info | |
| Image Data Resource (IDR) | A repository of image datasets from scientific publications | Bioimaging data Microbial biotechnology | Tool info Standards/Databases |
| OMERO | OMERO is an open-source client-server platform for managing, visualizing and analyzing microscopy images and associated metadata | Galaxy Bioimaging data | Tool info Training |

