What is the Molgenis tool assembly?
MOLGENIS is a modular web application for scientific data. MOLGENIS was born from molecular genetics research (and was called ‘molecular genetics information system’) but has become relevant to many other scientific areas such as biobanking, rare disease research, patient registries and even energy research. MOLGENIS provides user-friendly and scalable software infrastructures to capture, exchange, and exploit the large amounts of data that is being produced by scientific organizations all around the world. To get an idea of what the software can do, visit our MOLGENIS YouTube channel or our demo page via the related pages. MOLGENIS is an ELIXIR Recommended Interoperability Resource.
One of the key features is that it has a completely customisable data system, allowing you to model your data according to your needs. This creates flexibility that other, more static, database applications often lack. It is web-based, meaning you setup a server, install and configure MOLGENIS, load your data and share it. If your data is ready, setting up a useful online research database application can be done in few hours. Another key feature is that MOLGENIS is modular, having all kinds of extension modules to store and interact with your data. A good example are interfaces to create R and Python scripts that interact with your data. This enables you to add your own statistical modules to run statistical analysis, or create plots based on your data within the online environment.
Who can use the Molgenis tool assembly?
If you are a researcher, a (bio)informatician, a biomedical practitioner, a data manager or anyone else who handles a considerable amount of (scientific) data, then MOLGENIS is a software package that will help you in setting up an online database application in a short time, making your data query-able and allowing you to share your data with collaborators easily.The MOLGENIS software toolkit enables you to store, edit, analyse, and share your data efficiently.
MOLGENIS is open source, free to download and to install yourself. You can also purchase a MOLGENIS instance as a service ready to use from the Genomics Coordination Centrer (GCC), which is the main developer of MOLGENIS. GCC can also provide you with support on entering and managing your data-model on the servers.
For what purpose can Molgenis assembly be used?
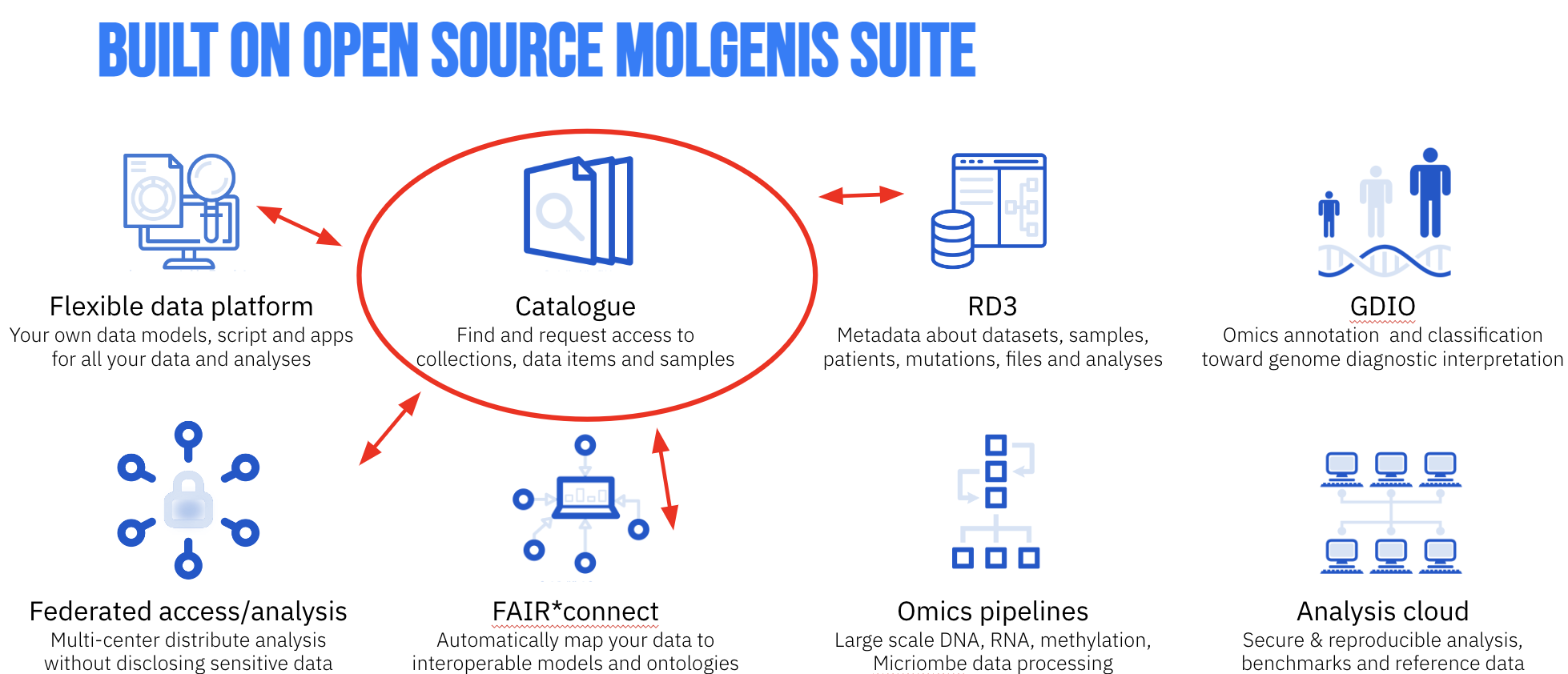
Figure 1. The Molgenis tool assembly.
Structured Data Management
Model, capture, and manage your data. Quickly upload data files, or enter data via user friendly forms. Refine your data model dynamically using MOLGENIS advanced ‘object-relational’ data definition format and the online metadata editor. Example: https://hfgp.bbmri.nl/
FAIR data sharing
Make your data findable, interoperable, accessible, reusable (FAIR). MOLGENIS aims to make data sharing and re-use should easy. MOLGENIS enables you to quickly create explorers for your data sets and variables to the outside world while preventing exposure of (sensitive) data values using the fine-grained permission system. Example: http://www.palgaopenbaredatabank.nl
Secure access
Easily control group, role and individual access. MOLGENIS data is organised following scientific practice. Data can be divided in research groups, within the groups you can assign roles such as ‘data manager’, ‘data editor’ and ‘data user’. Authentication can be ensured by connecting you institute account via SURFconext (NL) and BBMRI/LS Login (Europe) or using Google two-factor authentication.
Scripting & visualisation
Bioinformaticians can take full control in MOLGENIS. Add scripts in your favorite programming languages (e.g. R, javascript, python) and connect to the data using API’s to add great analysis tools and views. You can also create complete html + javascript apps to customize MOLGENIS further. Example: http://molgenis.org/ase Harmonization and integration A common task is to make your data interoperable and run combined analysis, which is much more powerful than running smaller analyses on each data set separately. MOLGENIS offers you multiple ‘FAIRification’ tools to find related data, codify your data contents and transform different tables into one standardized table. Examples: http://biobankconnect.org and http://molgenis.org/sorta
Task automation
Automate data upload, transformation and statistics. Frequently data from multiple sources must be combined for success. Therefore, data exchanges, transformations, and analyses must be repeated often. To enable in reproducible science, you can automate tasks with the MOLGENIS job scheduling tools. Questionnaires Get data directly from the source. Use the questionnaire tool to ask individuals for input. The tool provides chapters, subquestions, advanced validations, conditional or ‘skip’ questions and intermediate save (so you can fill in the rest of the survey later).
App development platform
Add your own user interfaces to the app store. MOLGENIS gives programmers the complete freedom to create HTML+JavaScript applications using MOLGENIS REST-style programmers interfaces. But it gets even better: you can upload these apps like plugins to become part of MOLGENIS itself and use them seamlessly.
High performance computing
Easily schedule large scale analysis jobs on a computer cluster. MOLGENIS does also provide a high performance computing framework. It is simply called ‘compute’ and it uses spreadsheets to define workflows, and templates to define workflow steps. It works on the HPC workload managers PBS and SLURM.
Related pages
More information
Training
Skip tool tableTools and resources on this page
| Tool or resource | Description | Related pages | Registry |
|---|---|---|---|
| MOLGENIS | Molgenis is a modular web application for scientific data. Molgenis provides researchers with user friendly and scalable software infrastructures to capture, exchange, and exploit the large amounts of data that is being produced by scientific organisations all around the world. | Identifiers | Tool info |

